Introduction
HQS Spectrum Tools is a software toolkit to simulate NMR spectra. It consists of hqs-nmr, containing simulation tools and hqs-nmr-parameters, providing NMR parameters for a variety of molecules. With HQS Spectrum Tools you can calculate and analyze NMR spectra for molecules or other spin systems.
Given the NMR parameters of a molecule, a simple python API can be used to calculate the associated NMR spectrum.
RUST and C++ components are used internally to achieve remarkable speed and accuracy, even for complex, large molecules.
Fastest NMR Spectrum Solver
Fastest NMR Spectrum Solver
“One of the fastest Hilbert-space NMR simulation tools I have ever seen, with a remarkably efficient frequency-domain implementation.” - Ilya Kuprov, Professor of Physics, University of Southampton
HQS Spectrum Tools is also an integral part of HQSpectrum, our end-to-end solution for NMR spectra analysis, which is available to you, if you have an existing HQStage account.
NMR spectra prediction is also a very promising use case for the quantum computer. To explore this possibility, we offer to extract the relevant spin Hamiltonians in struqture format for all molecules in the hqs-nmr-parameters database.
If you are interested in the theory behind NMR or the math of how to calculate NMR spectra, take a look at the background chapter.
Applications
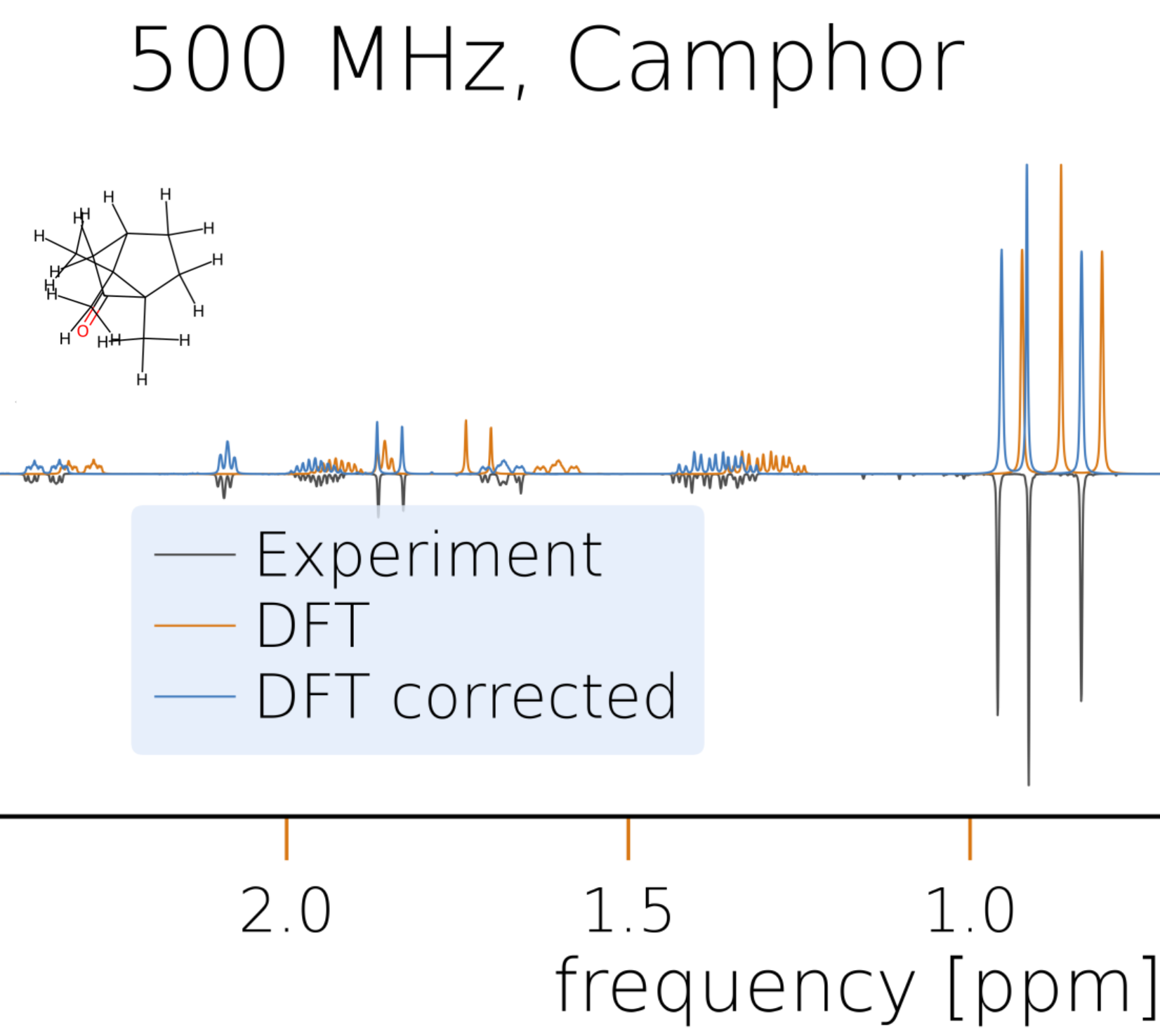
Nuclear magnetic resonance (NMR) spectroscopy is a key analytical tool in chemistry and related disciplines. Its broad application not only facilitates the identification of molecules, but also provides intricate details about their structure, dynamics, and chemical environment. The ability to compare an experimental NMR spectrum with theoretical predictions for various compounds is crucial for the accurate identification and characterization of chemical entities.
Our NMR spectrum solvers are designed for both speed and precision. They are based on a very efficient implementation in the frequency domain. With a variety of solver strategies at your disposal, you can leverage symmetries and clustering techniques either fully automated or customized according to your specifications. This flexibility allows you to tailor your approach based on the specific characteristics of your system, optimizing performance without sacrificing accuracy.
Getting started
To use HQS Spectrum Tools you need a running version of HQStage. You can check here how to set it up. Once you have HQStage correctly configured you can install HQS Spectrum Tools using the command:
hqstage install hqs-nmr
Please note that HQS Spectrum Tools is currently only supported on Linux.
For some of the functionalities of HQS Spectrum Tools (especially the provided example NMR parameters), the RDKit cheminformatics package is required. While packages are available on pypi.org, at present they are not officially supported by the RDKit development team. Therefore, it is strongly recommended to install it via the conda-forge channel to ensure the access to its latest version. The easiest way to install the library is running the following command in an active conda environment:
conda install rdkit
Note that RDKit will be installed automatically from pypi.org if it is not already present in the Python environment, for instance, if you do not have conda available.
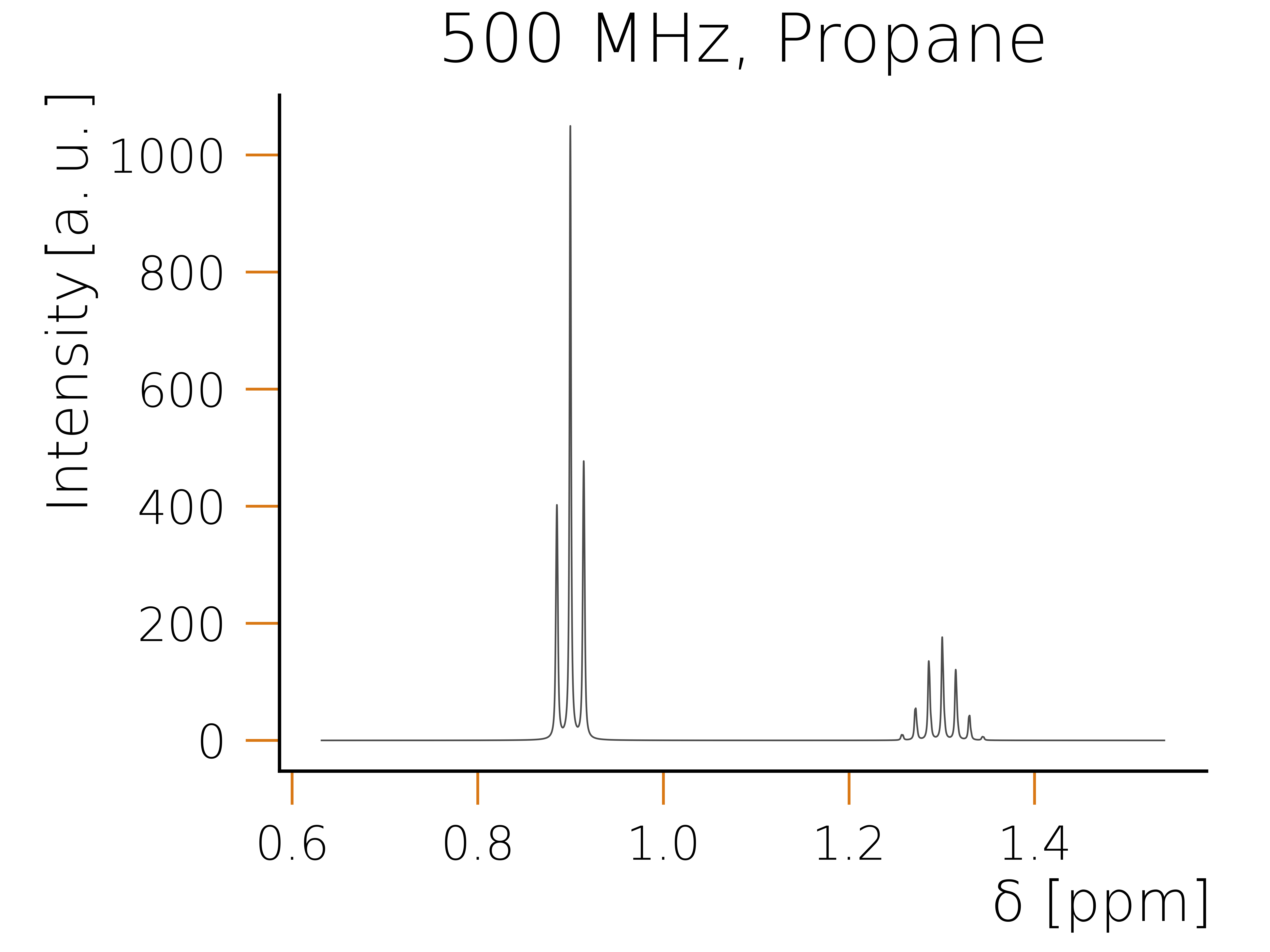
The following code snippet can be used to create your first program using HQS Spectrum Tools. For an expanded collection of examples please see the examples section.
from hqs_nmr.calculate import calculate_spectrum
from hqs_nmr.datatypes import NMRCalculationParameters
from hqs_nmr.visualization import plot_spectrum
from hqs_nmr_parameters.examples import molecules
from matplotlib import pyplot as plt
# Obtain example molecule of datatype NMRParameters.
molecule_parameters = molecules["C3H8"].spin_system()
# Define the calculation parameters. The only required field is the magnetic
# field in Tesla
calculation_parameters = NMRCalculationParameters(field_T=11.7433)
# Calculate the individual spin contributions of the spectrum.
nmr_result = calculate_spectrum(
molecule_parameters,
calculation_parameters
)
# Plot the spectrum.
plot_spectrum(nmr_result.spectrum)
plt.show()
Executing the code snippet using python should result in the following plot:
Features
- Frequency domain-based fast and precise NMR spectra calculation.
- Automatic implementation to compute accurate 1D NMR spectra with minimal truncation errors for large numbers of spins, featuring linear scaling with system size.
- NMR example parameters (chemical shifts and J-couplings) for molecules of different sizes, accessible via data classes containing structural information.
- Custom NMR parameters can be specified conveniently for NMR spectrum simulation.
- Easy construction of an NMR Spin Hamiltonian from provided or custom parameters using the
struqture-pypackage. - Extensive customization options of the NMR solver for expert users.
- Postprocessing methods to analyze simulated spectra and compare them to experiment.
You can get an overview of the HQS Spectrum Tools Python library in the components chapter or take a look at the API-documentation.