Local Usage
Prerequisites
- Python 3.10+ (use the
uvtabs to install Python on the fly) - macOS/Linux
- Account on https://cloud.quantumsimulations.de
- Get a free trial license at https://cloud.quantumsimulations.de/licenses
- Proceed with installation
Installation
While it’s best to start in a fresh virtual environment, you can simply pip install HQStage into any existing Python environment. Below we show both options and also our preferred way of using HQStage with uv.
pip install hqstage
hqstage init
The hqstage init command will prompt you to create an account and user token on https://cloud.quantumsimulations.de.
Intel’s Math Kernel Library
Intel’s Math Kernel Library
Some HQStage Modules use Intel’s MKL library to accelerate mathematical calculations. You can install MKL using
hqstage install mkl
The command ensures that runtime libraries of mkl are present in the correct format in your environments lib folder.
Free trial
To get started with your first quantum computing use case request a free trial. This free trial will add the HQS Spectrum Tools to your account which we will use to run our first example use case: a NMR spectrum prediction.
Running your first use case example: NMR
Install the requried modules
hqstage install ipython jupyter matplotlib mkl hqs_spectrum_tools
and open an ipython interpreter:
ipython
Now, copy paste the following code snippet:
from hqs_nmr_parameters import examples
from hqs_nmr import NMRCalculationParameters, calculate_spectrum
from matplotlib import pyplot as plt
from rdkit.Chem import Draw, MolFromMolBlock
print(f"Available moleucules: {examples.molecules.keys}")
molecule_name = '1,2,4-trichlorobenzene'
molecule_parameters = examples.molecules[molecule_name]
rdkit_mol = MolFromMolBlock(molecule_parameters.structures["Molfile"].content)
mol_image = Draw.MolToImage(rdkit_mol)
fig, ax = plt.subplots(figsize=(10, 6))
for field_T in [1, 5, 10]:
result = calculate_spectrum(molecule_parameters, NMRCalculationParameters(field_T=field_T))
ax.plot(result.spectrum.omegas_ppm, result.spectrum.intensity, label=f"B={field_T}T")
plt.legend()
ax.set_xlabel(r"$\delta$ [ppm]")
ax.set_ylabel("Intensity, arb. units")
ax.set_title(f"Spectrum of {molecule_name}")
ax_image = fig.add_axes(
[0.1,
0.6,
0.25,
0.25]
)
ax_image.imshow(mol_image)
ax_image.axis('off')
plt.show()
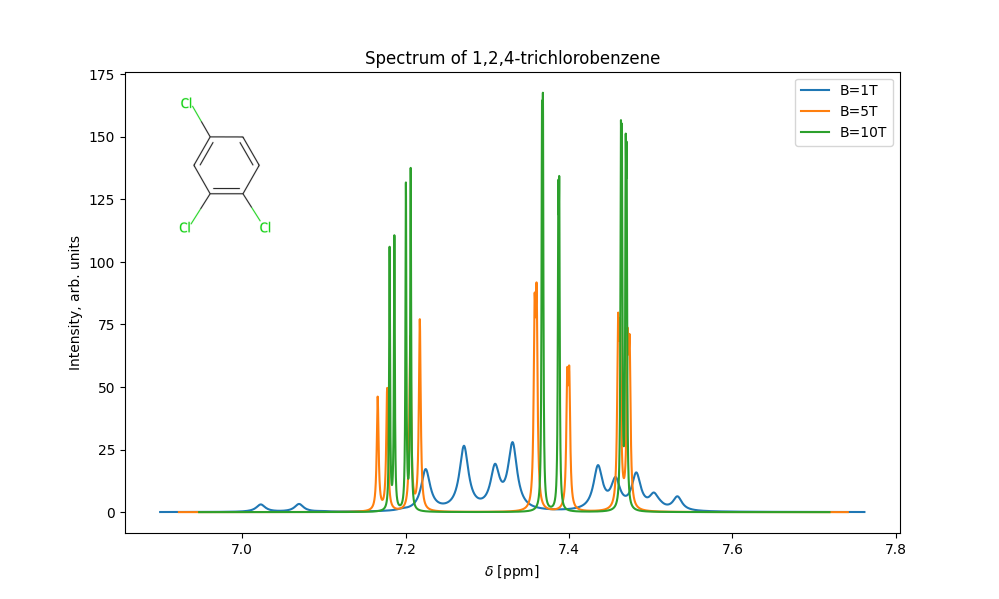
This will calculate the NMR spectrum for 1,2,4-trichlorobenzen and produce the following plot:
Downloading and using our examples
Download the use-case and spectrum examples
mkdir -p ~/HQStage/Examples
hqstage download-examples --download-dir ~/HQStage/Examples
jupyter lab --notebook-dir ~/HQStage/Examples
This will open jupyter lab in the folder where we have downladed the HQStage examples in. Fell free to explore the examples andadjust to your needs or continue reading more about our use cases